# 1. Prefix -------------------------------------------------------------------
# Remove all files from ls
rm(list = ls())
# Loading packages
require(survival)
require(flexsurv)
require(optimx)
require(numDeriv)
require(dplyr)
require(tibble)
require(car)
# 2. Loading dataset ----------------------------------------------------------
#Reading the example data set lung from the survival package
lung <- as.data.frame(survival::lung)
#Recode dichotomous vairables
lung$female <- ifelse(lung$sex == 2, 1, 0)
lung$status_n <- ifelse(lung$status == 2, 1, 0)In this blog post, we will fit a Poisson regression model by maximising its likelihood function using optimx() in R. As an example we will use the lung cancer data set included in the {survival} package. The data set includes information on 228 lung cancer patients from the North Central Cancer Treatment Group (NCCTG). Specifically, we will estimate the survival of lung cancer patients by sex and age using a simple Poisson regression model. You can download the code that I will use throughout post here. The general survival function \(S(t)\) for our model can be specified as
\[ S(t) = \exp(-\lambda t) \]
where the hazard function \(h(t)\) is equal to
\[ h(t) = \lambda = \exp(\alpha + \beta_1 x_{female} + \beta_2 x_{age}). \] To get started we first need to load all the packages that we will need for our estimations and set up the data set.
At this point we would usually call survreg() or flexsurvreg() to fit our Possion model. However, in this post we will use the likelihood function of our Possion regression model together with optimx() from the {optimx} package instead. For this we first need to find the likelihood function for our model and then use optimx() to find the values for our parameters, that maximise our likelihood function.
The log likelihood (\(\ln L_i\)) for a survival model can be specified as
\[ \ln L_i = d_i \ln h(t_i) + \ln S(t_i). \] Where \(d_i\) indicates whether the \(i^{th}\) subject experienced an event (1) or did not experience an event (0) during follow up and \(t_i\) is the time from the beginning of follow up until censoring.
To obtain the log likelihood function for our Possion model we can substitute \(h(t)\) and \(S(t)\) with our previously defined hazard and survival function respectively. Thus, we get the following equation for our log likelihood
\[\ln L_i = d_i \ln \lambda - \lambda t_i \]
where \(\lambda\) is defined as mentioned above.
The next step is now to write our likelihood function as a function in R, which can be maximised by optimx(). Please keep in mind, that optimx() by default minimises the function we pass to it. However, in our case we need to find the maximum of our likelihood function. To yield the estimates, that maximise our function we can just ask optimx() to minimise the negative of our likelihood. For more information on setting up the likelihood function for optimx() or optim() please take a look at this earlier blog post.
Lets set up our likelihood function in R.
# 3. Define log-likelihood function for Poisson regression model --------------
negll <- function(par){
#Extract guesses for alpha, beta1 and beta2
alpha <- par[1]
beta1 <- par[2]
beta2 <- par[3]
#Define dependent and independent variables
t <- lung$time
d <- lung$status_n
x1 <- lung$female
x2 <- lung$age
#Calculate lambda
lambda <- exp(alpha + beta1 * x1 + beta2 * x2)
#Estimate negetive log likelihood value
val <- -sum(d * log(lambda) - lambda * t)
return(val)
}To improve the optimisation we can further pass the gradient function of our likelihood function to our optimx() call. After partially deriving \(L_i\) for \(\alpha\) and \(\beta_i\) we yield the two following equations for the gradient of \(L_i\).
\[ \sum d_i - \lambda_i t_i = 0\]
\[ \sum d_i x_{ij} - \lambda_i x_{ij} t = 0\]
Given these gradient equations we can now define our gradient function in R. For this we need to create a function, that returns the gradient for each of our unknown parameters. Since we have three unknown parameters our gradient function will return a vector gg with three values.
# 4. Define gradient function for Poisson regression model -------------------
negll.grad <- function(par){
#Extract guesses for alpha and beta1
alpha <- par[1]
beta1 <- par[2]
beta2 <- par[3]
#Define dependent and independent variables
t <- lung$time
d <- lung$status_n
x1 <- lung$female
x2 <- lung$age
#Create output vector
n <- length(par[1])
gg <- as.vector(rep(0, n))
#Calculate pi and xb
lambda <- exp(alpha + beta1 * x1 + beta2 * x2)
#Calculate gradients for alpha and beta1
gg[1] <- -sum(d - lambda * t)
gg[2] <- -sum(d * x1 - lambda * x1 * t)
gg[3] <- -sum(d * x2 - lambda * x2 * t)
return(gg)
}We can compare the results of our gradient function with the results from the grad() function included in the {numDeriv} package, before we begin with the optimisation of our functions. This is just a check to be sure our gradient function works properly.
# 4.1 Compare gradient function with numeric approximation of gradient ========
# compare gradient at 0, 0, 0
mygrad <- negll.grad(c(0, 0, 0))
numgrad <- grad(x = c(0, 0, 0), func = negll)
all.equal(mygrad, numgrad)[1] TRUELooks like our gradient functions does a good job. Now that we have all the functions and information we need for our optimisation, we can call optimx() and pass our functions to it.
The output of optimx() provides us with estimates for our coefficients and information regarding whether the optimisation algorithm converged (convcode == 0) besides the maximum value of the negative log likelihood obtained by the different algorithms. Hence, it is useful to sort the results by convcode and value.
# 5. Find maximum of log-likelihood function ----------------------------------
# Passing names to the values in the par vector improves readability of results
opt <- optimx(par = c(alpha = 0, beta_female = 0, beta_age = 0),
fn = negll,
gr = negll.grad,
hessian = TRUE,
control = list(trace = 0, all.methods = TRUE))
# Show results for optimisation alogrithms, that convergered (convcode == 0)
summary(opt, order = "value") %>%
rownames_to_column("algorithm") %>%
filter(convcode == 0) %>%
select(algorithm, alpha, beta_female, beta_age, value) algorithm alpha beta_female beta_age value
1 nlminb -6.840606 -0.4809343 0.01561870 1156.099
2 BFGS -6.840627 -0.4809436 0.01561907 1156.099
3 L-BFGS-B -6.840636 -0.4809316 0.01561902 1156.099
4 Nelder-Mead -6.832428 -0.4814582 0.01547911 1156.099The summary of our optimx() call shows, that the nlminb algorithm yielded the best result. Lets see if this result is equal to the results we will get, if we use flexsurvreg from the {flexsurv} package to fit our desired model.
# 6. Estimate regression coeficents using flexsurvreg -------------------------
pois_model <- flexsurvreg(Surv(time, status_n == 1) ~ female + age,
data = lung,
dist = "exp")
# 7. Comparing results from optimx and flexsurvreg ----------------------------
pois_results <- unname(coef(pois_model))
coef_opt <- coef(opt)
lapply(1:nrow(coef_opt), function(i){
opt_name <- attributes(coef_opt)$dimnames[[1]][i]
mle_pois1 <- (coef_opt[i, 1] - pois_results[1])
mle_pois2 <- (coef_opt[i, 2] - pois_results[2])
mle_pois3 <- (coef_opt[i, 3] - pois_results[3])
mean_dif <- mean(mle_pois1, mle_pois2, mle_pois3, na.rm = TRUE)
data.frame(opt_name, mean_dif)
}) %>%
bind_rows() %>%
filter(!is.na(mean_dif)) %>%
mutate(mean_dif = abs(mean_dif)) %>%
arrange(mean_dif) opt_name mean_dif
1 nlminb 2.678650e-07
2 BFGS 2.091779e-05
3 L-BFGS-B 2.911668e-05
4 Nelder-Mead 8.178948e-03
5 CG 6.816256e+00
6 Rvmmin 6.840606e+00The mean difference between our estimates and the estimates obtained by using flexsurvreg() are close to zero. Seems like our optimisation using the log likelihood did a good job.
However, the result obtained with flexsurvreg() provided us with estimates for the standard errors (SEs) of our hazard estimates, too. Since the measurement of uncertainty is at the heart of statistics, I think it is worthwhile to obtain the SEs for our estimates with the information provided by our optimx() call. For a more detailed discussion on how this is done please take a look at one of my previous blog posts here.
Let’s obtain the SEs for our model by using the results from our optimx() call and compare them with the SEs obtained by flexsurvreg().
# 8. Estimate the standard error ----------------------------------------------
#Extract hessian matrix for nlminb optimisation
hessian_m <- attributes(opt)$details["nlminb", "nhatend"][[1]]
# Estimate SE based on hession matrix
fisher_info <- solve(hessian_m)
prop_se <- sqrt(diag(fisher_info))
# Compare the estimated SE from our model with the one from the flexsurv model
# Note use res.t to get the estimates on the reale scale without transformaiton
ses <- data.frame(se_nlminb = prop_se,
se_felxsurvreg = pois_model$res.t[, "se"]) %>%
print() se_nlminb se_felxsurvreg
rate 0.587477415 0.58747740
female 0.167094278 0.16709429
age 0.009105681 0.00910568all.equal(ses[, "se_nlminb"], ses[, "se_felxsurvreg"])[1] "Mean relative difference: 3.697145e-08"Looks like we got nearly equal results. Let us use these information and estimate the 95% confidence intervals (CIs) for our estimates now.
# 9. Estimate 95%CIs using estimation of SE -----------------------------------
# Extracting estimates from nlminb optimisaiton
coef_test <- coef(opt)["nlminb",]
# Compute 95%CIs
upper <- coef_test + 1.96 * prop_se
lower <- coef_test - 1.96 * prop_se
# Print 95%CIs
data.frame(Estimate = coef_test,
CI_lower = lower,
CI_upper = upper,
se = prop_se) Estimate CI_lower CI_upper se
alpha -6.8406062 -7.992061931 -5.68915046 0.587477415
beta_female -0.4809343 -0.808439062 -0.15342949 0.167094278
beta_age 0.0156187 -0.002228433 0.03346584 0.009105681One usual way to plot the results of our estimation is plotting the survival function \(S(t)\). Since, uncertainty is important I also want to plot the CI for our survival function. To obtain estimates for the SE of the survival function \(S(t)\) is a little bit more complicated. However, the amazing deltaMethod() function included in the {car} package makes it fairly easy to obtain estimates for the SEs. We just need to provide deltaMethod() with a vector of our coefficients, our covariance matrix and the computation for which we would like to obtain the SEs.
# 10. Plot survival curve with 95%-CI -----------------------------------------
# 10.1 Use Delta Method to compute CIs across time of follow-up ===============
# Get coefficents for nlminb optimisation
nlminb_coef <- coef(opt)["nlminb", ]
# Compute CIs for a 60 year old female across follow-up time
surv_optim_female <- lapply(as.list(seq(0.01, 1000.01, 10)), function(t){
g <- paste("exp(-exp(alpha + beta_female + 60 * beta_age) *", t, ")")
fit <- deltaMethod(nlminb_coef, g, solve(hessian_m))
data.frame(time = t,
estimate = fit[, "Estimate"],
ci_low = fit[, "2.5 %"],
ci_up = fit[, "97.5 %"])
}) %>%
bind_rows()We can now use these information to plot our survival curve \(S(t)\) together with a grey shaded area that indicates the CIs for our survival function.
# 10.2 Plot survival curve with CIs ===========================================
plot(surv_optim_female$time,
surv_optim_female$estimate,
ylim = c(0, 1),
type = "n",
xlab = "Time in Days",
ylab = "S(t)",
main = "Survival after lung cancer \n for 60 year old females")
polygon(c(surv_optim_female$time, rev(surv_optim_female$time)),
c(surv_optim_female$ci_low, rev(surv_optim_female$ci_up)),
border = NA,
col = "grey")
lines(surv_optim_female$time,
surv_optim_female$estimate)
legend(0, 0.15,
fill = "grey",
"95% CI")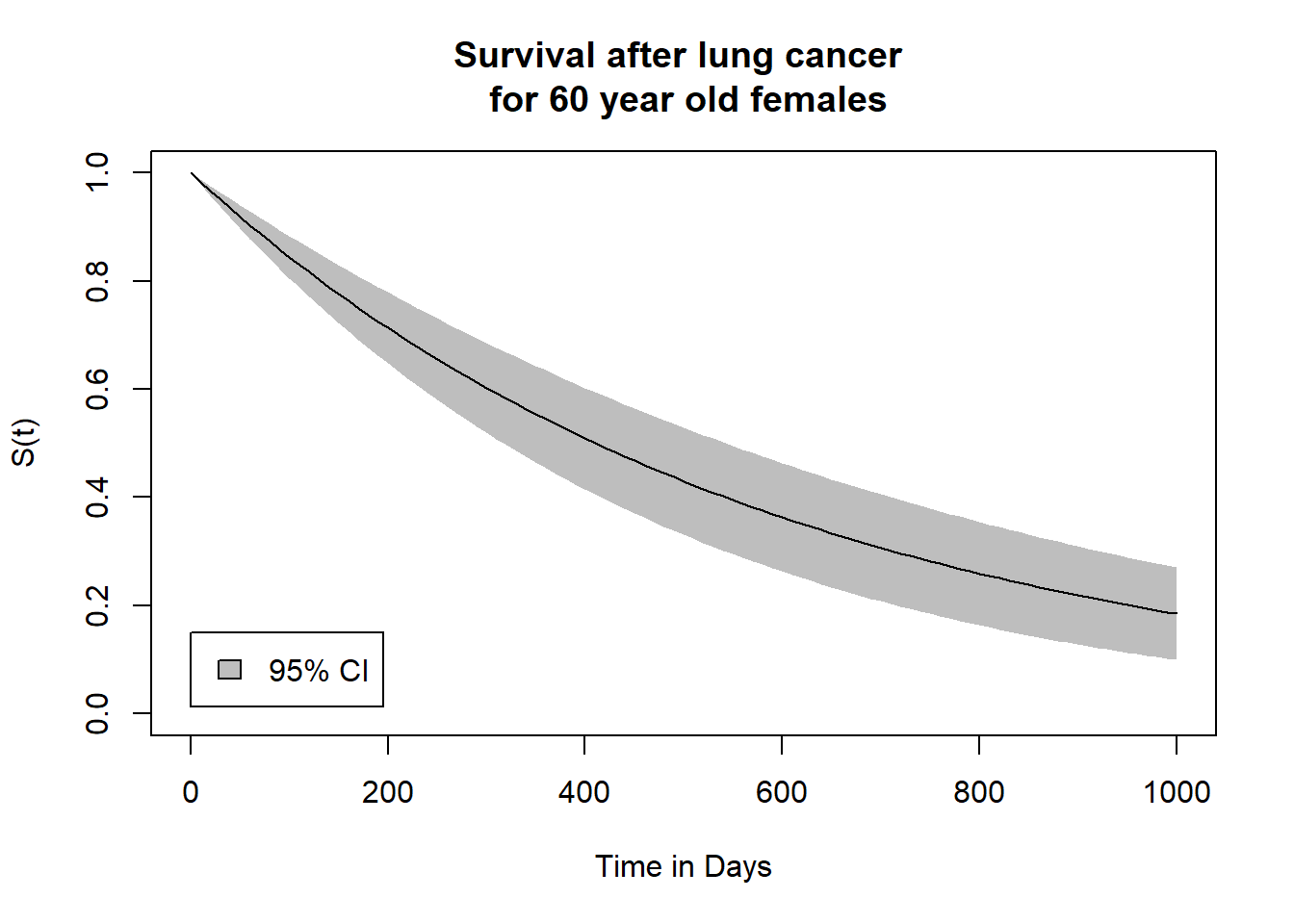
To sum it up, in this blog post we learned how to fit a Possion regression model using the log likelihood function in R instead of going the usual way of calling survreg() or flexsurvreg(). I think doing this is a good way of gaining a deeper understanding of how estimates for regression models are obtained. In my next post I will take this a step further and show how we can fit a Weibull regression model in R using the log likelihood function in combination with optimx().